Pre-regionalization using polynomial mapping#
Here, we aim to employ some physiographic descriptors to find the pre-regionalization mapping using polynomial functions. Six descriptors are considered in this example, which are:
Slope
Drainage density
Karst
Woodland
Urban
Soil water storage
First, open a Python interface:
python3
Imports#
In [1]: import smash
In [2]: import matplotlib.pyplot as plt
Model object creation#
To perform the calibrations, you need to create a smash.Model object.
For this case, we will use the Lez dataset.
Load the setup and mesh dictionaries using the smash.load_dataset() method and create the smash.Model object.
In [3]: setup, mesh = smash.load_dataset("Lez")
In [4]: model = smash.Model(setup, mesh)
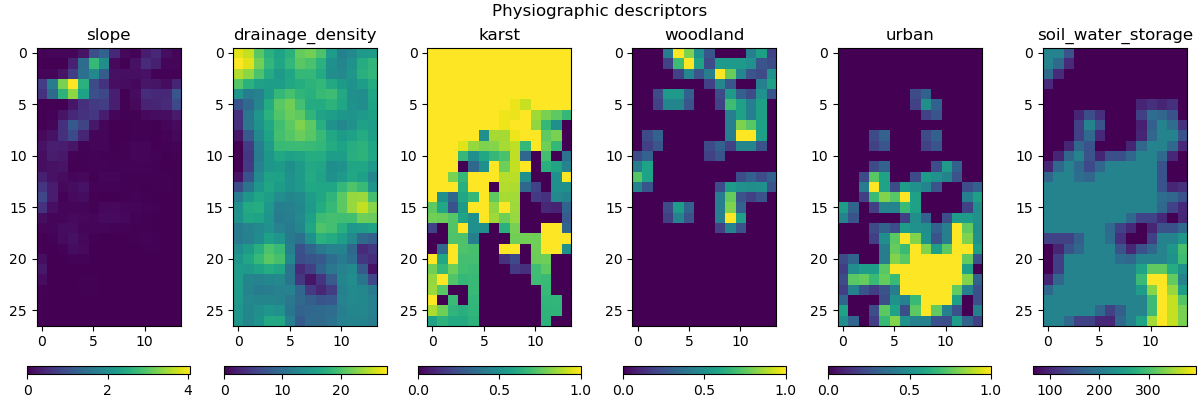
Visualization of descriptors#
This method requires input descriptors, which were provided during the creation of the Model object. We can visualize these descriptors and verify if they were successfully loaded:
In [5]: model.input_data.descriptor.shape
Out[5]: (27, 14, 6)
In [6]: desc_name = model.setup.descriptor_name
In [7]: fig, axes = plt.subplots(1, len(desc_name), figsize=(12,4), constrained_layout=True)
In [8]: for i, ax in enumerate(axes):
...: ax.set_title(desc_name[i]);
...: im = ax.imshow(model.input_data.descriptor[..., i]);
...: cbar = fig.colorbar(im, ax=ax, orientation="horizontal");
...: cbar.ax.tick_params();
...:
In [9]: fig.suptitle("Physiographic descriptors");
# Reset figsize to the Matplotlib default
In [10]: plt.figure(figsize=plt.rcParamsDefault['figure.figsize']);
Finding a uniform first guess#
Similar to the fully-distributed optimization method, providing a uniform first guess is recommended for this method. In this case, we use the \(\mathrm{SBS}\) algorithm to find such a first guess:
In [11]: model_su = model.optimize(mapping="uniform", algorithm="sbs", options={"maxiter": 2});
Hint
You may want to refer to the Bayesian estimation section for information on how to improve the first guess using a Bayesian estimation approach.
Optimizing hyperparameters for pre-regionalization mapping#
There are two types of polynomial mapping that can be employed for pre-regionalization:
hyper-linear: a linear mapping where the hyperparameters to be estimated are the coefficients.hyper-polynomial: a polynomial mapping where the hyperparameters to be estimated are the coefficients and the degree.
As an example, the hyper-polynomial mapping can be combined with the variational calibration algorithm \(\mathrm{L}\text{-}\mathrm{BFGS}\text{-}\mathrm{B}\) as shown below:
In [12]: model_hp = model_su.optimize(
....: mapping="hyper-polynomial",
....: algorithm="l-bfgs-b",
....: options={"maxiter": 30}
....: )
....:
Some information are also provided during the optimization:
</> Optimize Model
Mapping: 'hyper-polynomial' k(x) = a0 + a1 * D1 ** b1 + ... + an * Dn ** bn
Algorithm: 'l-bfgs-b'
Jobs function: [ nse ]
wJobs: [ 1.0 ]
Jreg function: 'prior'
wJreg: 0.000000
Nx: 1
Np: 52 [ cp cft exc lr ]
Ns: 0 [ ]
Ng: 1 [ Y3204040 ]
wg: 1 [ 1.0 ]
At iterate 0 nfg = 1 J = 0.176090 |proj g| = 0.000000
At iterate 1 nfg = 3 J = 0.174870 |proj g| = 0.160574
At iterate 2 nfg = 4 J = 0.173283 |proj g| = 0.059085
At iterate 3 nfg = 5 J = 0.172243 |proj g| = 0.043317
At iterate 4 nfg = 6 J = 0.171181 |proj g| = 0.045926
At iterate 5 nfg = 7 J = 0.170460 |proj g| = 0.023084
At iterate 6 nfg = 8 J = 0.169568 |proj g| = 0.025826
At iterate 7 nfg = 9 J = 0.168186 |proj g| = 0.046616
At iterate 8 nfg = 10 J = 0.165931 |proj g| = 0.069842
At iterate 9 nfg = 11 J = 0.160961 |proj g| = 0.077036
At iterate 10 nfg = 13 J = 0.152905 |proj g| = 0.209049
At iterate 11 nfg = 14 J = 0.148905 |proj g| = 0.056703
...
At iterate 28 nfg = 34 J = 0.133811 |proj g| = 0.008069
At iterate 29 nfg = 35 J = 0.133753 |proj g| = 0.003290
At iterate 30 nfg = 36 J = 0.133749 |proj g| = 0.001082
STOP: TOTAL NO. OF ITERATION EXCEEDS LIMIT
Visualization of results#
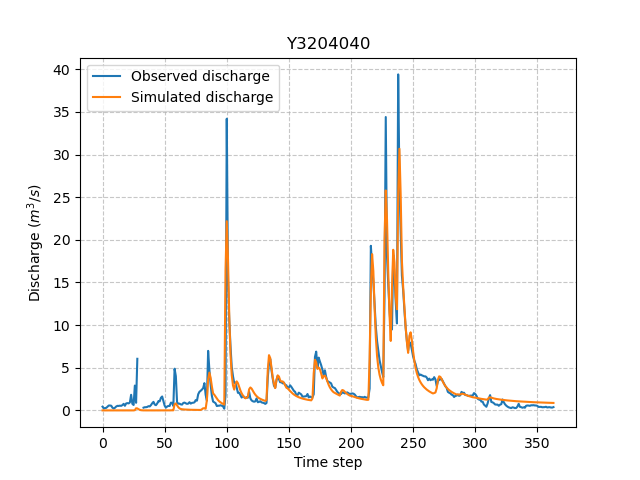
Now we can visualize the simulated discharge:
In [13]: qo = model_hp.input_data.qobs[0,:].copy()
In [14]: qo = np.where(qo<0, np.nan, qo) # to deal with missing data
In [15]: plt.plot(qo, label="Observed discharge");
In [16]: plt.plot(model_hp.output.qsim[0,:], label="Simulated discharge");
In [17]: plt.grid(alpha=.7, ls="--");
In [18]: plt.xlabel("Time step");
In [19]: plt.ylabel("Discharge $(m^3/s)$");
In [20]: plt.title(model_hp.mesh.code[0]);
In [21]: plt.legend();
The cost value:
In [22]: model_hp.output.cost
Out[22]: 0.13374879956245422
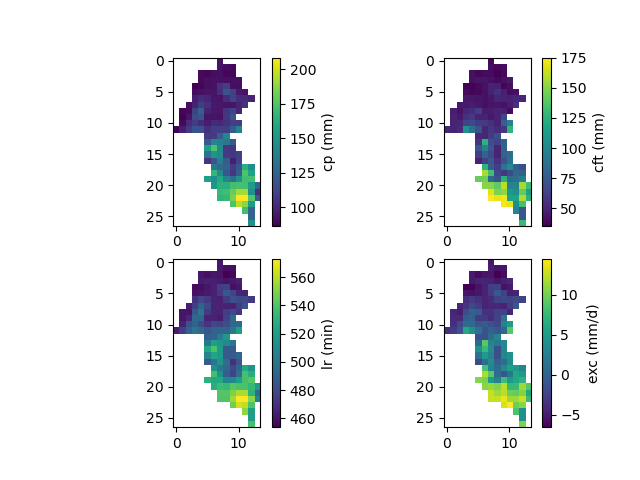
And finally, the spatially distributed model parameters constrained by physiographic descriptors:
In [23]: ma = (model_hp.mesh.active_cell == 0)
In [24]: ma_cp = np.where(ma, np.nan, model_hp.parameters.cp)
In [25]: ma_cft = np.where(ma, np.nan, model_hp.parameters.cft)
In [26]: ma_lr = np.where(ma, np.nan, model_hp.parameters.lr)
In [27]: ma_exc = np.where(ma, np.nan, model_hp.parameters.exc)
In [28]: f, ax = plt.subplots(2, 2)
In [29]: map_cp = ax[0,0].imshow(ma_cp);
In [30]: f.colorbar(map_cp, ax=ax[0,0], label="cp (mm)");
In [31]: map_cft = ax[0,1].imshow(ma_cft);
In [32]: f.colorbar(map_cft, ax=ax[0,1], label="cft (mm)");
In [33]: map_lr = ax[1,0].imshow(ma_lr);
In [34]: f.colorbar(map_lr, ax=ax[1,0], label="lr (min)");
In [35]: map_exc = ax[1,1].imshow(ma_exc);
In [36]: f.colorbar(map_exc, ax=ax[1,1], label="exc (mm/d)");